
Application Note
Detection of autophagy using automated imaging
- Quantify compound effects on the process of autophagy
- Detect and characterize small objects while using the nuclear marker to segment cells
- Ensure reliable statistical results using automated imaging
Introduction
Oksana Sirenko | Senior Research Scientist | Molecular Devices
Autophagy as a promising target for drug therapies
Autophagy and its dysregulation has been found to play an important role in neurodegenerative diseases and cancers, therefore the discovery of novel therapeutic targets along this process has emerged as a promising approach for drug therapies. Autophagy is a regulated process of degrading and recycling damaged proteins and organelles in response to cellular stress.1,2 The vesicle called the autophagosome is assembled by the formation of a double membrane around the cellular component marked for destruction.1 The autophagosome vesicle then fuses with the lysosome to deliver its contents for degradation by lysosomal hydrolases.
Autophagy has a variety of complex physiological and pathophysiological roles, such as adaptation to nutrient starvation, clearance of damaged intracellular proteins and organelles, cell development, antiaging, elimination of microorganisms, cell death, tumor suppression, and antigen presentation.2 Mitophagy is the selective degradation of mitochondria by autophagy.3 This process often eliminates defective mitochondria following cellular damage or stress.
Materials
- PC12 human neuroblastoma cell line (ATCC)
- 384-well plates (Greiner)
- Cyto-ID ® Autophagy Detection Kit (ENZO Life Sciences)
- Orange Mitochondrial Integrity Dye
- Hoechst 33342
- ImageXpress ® Pico Automated Cell Imaging System (Molecular Devices)
- CellReporterXpress ™ Image Acquisition and Analysis Software (Molecular Devices)
Detect and quantify autophagosomal particles using automated cell imaging
Evaluate effects on autophagy
Here, we evaluated the efficiency of the ImageXpress Pico Automated Cell Imaging System for detection and quantitation of autophagosomal particles. The PC12 human neuroblastoma cell line was used as a model for assay development. Cells were plated into 384-well plates at 6,000 cells/ well and incubated for 48 h. To evaluate effects on autophagy, cells were treated with various concentrations of chloroquine or verapamil for 48 h. Verapamil induces autophagy while chloroquine inhibits degradation of autophagosomes that lead to accumulation of particles. After treatment, live cells were stained with the CYTO-ID® Autophagy Detection Kit for tracking autophagosomes. In addition, we used a fluorescent mitochondrial dye for the detection of mitochondria and Hoechst to identify nuclei (0.2 μM and 1 μM, respectively). Images were taken using the ImageXpress Pico system with 20X or 40X magnification and three detection channels for the appropriate dyes (FITC, TRITC, and DAPI, respectively). One image per well was captured at 20X magnification, while 2–4 images were acquired per well at 40X magnification to ensure reliable statistical results. Images were analyzed using the Autophagy or Mitochondria application protocols in CellReporterXpress Image Acquisition and Analysis Software.
The algorithms detect and characterize small objects, such as autophagosomes or mitochondria in the cytoplasm of cells, while using the nuclear marker to segment cells. Quantitative measurements for detection of autophagy include total or average numbers of granules, the total or average areas of granules (subcellular objects), as well as intensity measurements. Representative images and analysis masks for cells treated with chloroquine (30 μM) are shown in the Figure 1. Concentration responses for chloroquine and verapamil were examined and EC50 values were evaluated. Chloroquine and verapamil treatment increased autophagy levels by 4.6 and 3.3-fold, respectively, as determined by total autophagosome counts (EC50 values were 4.0 and 3.6 μM respectively).
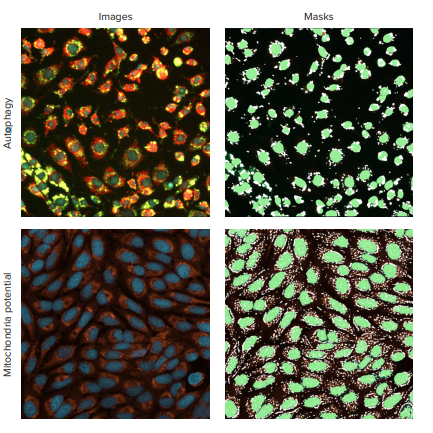
Figure 1. Detection of autophagosomes. The top panel shows representative autophagy cells image and analysis mask for PC12 cells treated with an autophagy inducer chloroquine (30 μM) and stained with Cyto-ID dye. In the CellReporterXpress software, Spot Segmentation was used to identify autophagy granules. Nuclei are shown in blue, mitochondria orange, autophagy in green. The lower panel shows an image and analysis mask for mitochondria detection. The untreated cells were stained with the orange mitochondrial integrity dye.
Conclusion
The method along with the CYTO-ID dye allows detection of autophagy particles and can be used for assay development and quantitation of the compound effects on the process of autophagy.
Reference
- Mizushima, N; Komatsu, M (2011). Autophagy: renovation of cells and tissues. Cell. 147: 728–41
- Mizushima, N; (2007). Autophagy: process and function, Genes & Dev. 21: 2861-2873
- Lemasters, J (2005). Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Research. 8: 3–5