
Application Note
Sensitive fluorescent quantitation of DNA with the Quant-iT PicoGreen dsDNA Assay Kit
- Sensitive fluorescent quantitation of DNA down to 50 pg/mL
- Linear dynamic range spanning over four orders of magnitude
- Easy analysis of results with preconfigured protocol in SoftMax Pro Software
Joyce Itatani | Application Scientist | Molecular Devices
Cathy Olsen | Sr. Application Scientist | Molecular Devices
Introduction
Double-stranded DNA is typically quantitated in microplate readers by measuring the absorbance of the DNA solution at 260 nm. However, this method is only able to measure down to about 250 ng/mL on a typical absorbance microplate reader. For biological applications involving small samples, such as next-generation sequencing and quantitation of DNA amplification products, more sensitive methods are needed. The Quant-iT PicoGreen dsDNA Assay Kit from Thermo Fisher Scientific is more specific for DNA and is about 1000 times more sensitive than traditional absorbance methods. The dynamic range of this assay in microplate format, as stated in the product manual, is from 250 pg/mL to 1000 ng/mL with a single dye concentration. Here, we demonstrate that with Molecular Devices SpectraMax® microplate readers and the Quant-iT PicoGreen assay, users can reliably measure concentrations as low as 50 pg/mL of double-stranded DNA.
To maximize sensitivity of the assay, it is necessary to use optimal excitation and emission wavelengths. Unlike filter-based plate readers, the dual monochromators in the SpectraMax iD5 Multi-Mode Microplate Reader and other SpectraMax readers allow the selection of any wavelength within the reader’s stated range. It is important to determine the excitation and emission wavelengths that provide the best sensitivity and dynamic range for the assay, since these may differ somewhat from those used with filter-based plate readers, and from wavelengths recommended in the kit manual.
Materials
- Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific cat. #P7589)
- 96-well solid black microplate (Greiner Bio-One cat. #655076)
- Light Safe Black Microtubes (Argos cat. #T7100BK)
- Microplate readers (note: additional SpectraMax readers not listed here have similar performance for the PicoGreen assay)
- SpectraMax Mini Multi-Mode Microplate Reader (Molecular Devices P/N SMAX MINI AF), with fluorescence filter cube FL-535 (Molecular Devices P/N 5089097)
- SpectraMax® iD5 Multi-Mode Microplate Reader (Molecular Devices P/N #iD5)
- SpectraMax® i3x Multi-Mode Microplate Reader (Molecular Devices P/N #i3x)
- SpectraMax® M5 Multi-Mode Microplate Reader (Molecular Devices P/N #M5)
- SpectraMax® Gemini™ EM Microplate Reader (Molecular Devices P/N #EM)
Methods
Instrument and protocol setup
- Turn on the microplate reader.
- Launch SoftMax® Pro Software and open the PicoGreen fluorescence protocol from the Protocols dropdown menu. Depending on which SpectraMax reader you are using, you may need to enter the optimized settings for the assay (see Table 1).
- Select Wells to Read and Assay Plate Type by clicking on “Settings” and locating the options on the left side of the screen.
- Click the Template button to open a window where you can assign wells of the microplate to pre-set template groups. Use the dropdown menu to select the appropriate template group. There are preconfigured template groups in the PicoGreen fluorescence protocol including Standards, Unknowns, and Unknowns_NoDiln (use for samples that are undiluted). Assigning wells to preset template groups populates group tables in the protocol with the corresponding data that is acquired when the microplate is read.
Prepare the assay
The method for this assay follows the instructions in the product information sheet for QuantiT PicoGreen dsDNA Reagent and Kits, except the assay volume is proportionately reduced from 2.0 mL to 200 μL to fit a 96-well microplate format.
- Prepare 1X TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) by diluting the concentrated buffer from the kit 20-fold with distilled DNase-free water.
- Prepare an aqueous working solution of Quant-iT PicoGreen reagent by making a 200-fold dilution of the concentrated DMSO solution in TE buffer (prepared above). Preparation of the solution in a plastic container, rather than glass, is recommended, as the reagent may adsorb to glass surfaces. Protect the solution from light by using brown or black tubes, or by covering with foil. This solution should be used within a few hours of its preparation.
- DNA standard curve: Prepare a 2 μg/mL stock solution of dsDNA in TE. The lambda DNA standard provided with the kit can be diluted 50-fold in TE to make the 2 μg/mL solution.
Note: In some cases, it may be preferable to make the standard curve using DNA similar to the type being assayed.
Emission: 535 nm
bandwidth 9 nm
Emission: 525 nm,
bandwidth 15 nm
Emission: 525 nm,
Emission cutoff: 515 nm
Emission: 525 nm,
Emission cutoff: 515 nm
Integration time: 200 ms
Read height: Optimize for microplate used
Flashes per read: 10
Read height: Optimize for microplate used
Flashes per read: 10
Flashes per read: 10
Integration time: 400 ms
Read height: Optimize for microplate used
Table 1. Instrument settings for SpectraMax readers. For SpectraMax iD5, i3x, and Mini readers, the read height setting should be optimized for the microplate used. Note: Additional readers with similar performance are listed.
- If desired, a high-range standard curve may be prepared from 1 ng/mL to 1 μg/mL, or a low-range standard curve may be prepared from 25 pg/mL to 25 ng/mL. For high-range or low-range curves, 1:10 dilutions may be used. For the low-range curve, dilute the 2 μg/mL solution 40-fold to yield a 50 ng/mL starting solution.
For this application note, a series of standards ranging from 50 pg/mL to 1 μg/mL was set up as a 1:3 dilution series. - Pipette standards into a solid black 96-well microplate at 100 μL per well, preferably in triplicate. Be sure to include a set of buffer blank wells containing TE only (no DNA).
- Add 100 μL of the aqueous working solution of QuantiT PicoGreen reagent to each well (this results in a 1:2 dilution of the standard already in the wells). Mix well by trituration or plate shaker and incubate for 2 to 5 minutes at room temperature, protected from light.
Read the microplate
- If using a SpectraMax M Series reader, make sure the purple plate adapter is in the microplate reader drawer. Place the microplate in the drawer.
- Click the Read button in SoftMax Pro Software.
Analyze the data
- After the microplate has been read, the relative fluorescence units (RFUs) will be displayed in the Plate section. The data will be analyzed in the Group Tables that were created when the template was set up. For an example of representative data from a Group Table, see Table 2.
- Standards assigned in the Template (and thus displayed in the Standards group table) will be automatically plotted in the Standard Curve section of the protocol. A linear curve fit is applied by default, but a log-log fit may be used when plotting a standard curve over a wide dynamic range. Curve fits are chosen from the dropdown Fit menu in the graph section.
Table 2. DNA standards.
Results
DNA standards ranging from 50 pg/mL to 1 μg/mL were detected using the Quant-iT PicoGreen dsDNA Assay Kit and SpectraMax readers (data from the SpectraMax iD5 reader are shown, but other SpectraMax readers gave similar results). SoftMax Pro Software automatically calculated average RFU, standard deviation, and %CV for each set of standard replicates. A standard curve was plotted using the log-log curve fit in SoftMax Pro Software (Figure 1). Sensitivity down to 50 pg/mL was observed using the 96-well microplate format and standard limit of detection calculation of three times standard deviation of the blank. This is well below the lower limit of 250 pg/ mL stated in the Quant-iT PicoGreen assay product insert. Figure 2 shows the high-range (A) and low-range (B) standard curves. Linearity was excellent throughout the standards’ range (r2 ≥ 0.99 for each curve shown).
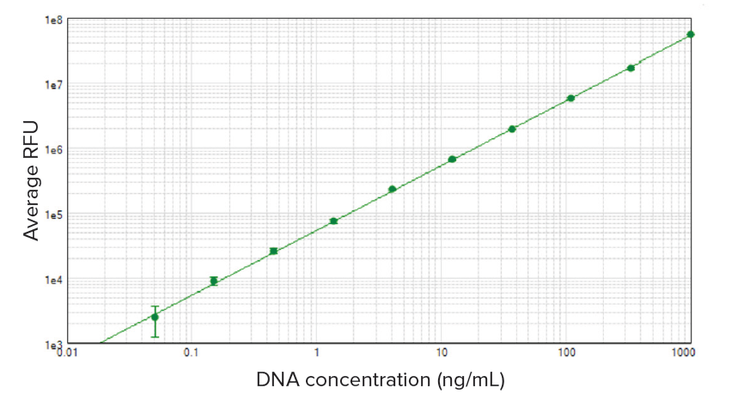
Figure 1. DNA standard curve. DNA standards ranging from 50 pg/mL to 1000 ng/mL were assayed using the Quant-iT PicoGreen dsDNA Assay Kit on the SpectraMax iD5 reader. The standard curve was plotted using the log-log curve fit in SoftMax Pro Software (r2 = 1.00).
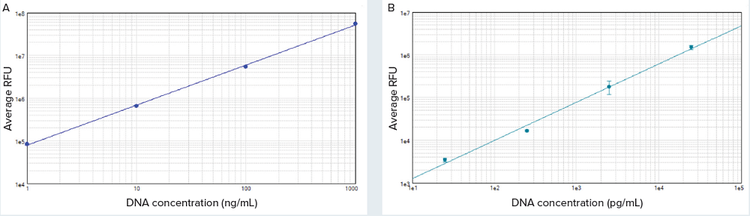
Figure 2. High-range (A) and low-range (B) standard curves. Curves were plotted using the log-log curve fit in SoftMax Pro Software (both curves, r2 = 0.99).
Conclusion
The Quant-iT PicoGreen dsDNA Assay Kit, when run on a SpectraMax microplate reader with SoftMax Pro Software, is a quick, sensitive detection method for double-stranded DNA. The analysis capabilities of the software provide quantitation in an easy-to-read, user customizable report format. A preconfigured protocol is available in the software to facilitate rapid assay setup.
Joyce Itatani | Application Scientist | Molecular Devices
Cathy Olsen | Sr. Application Scientist | Molecular Devices
简介
双链 DNA 通常通过测定 DNA 溶液在 260 nm 处的吸光度在 酶标仪中进行定量。然而,该方法在典型的光吸收酶标仪上只 能测量到约 250 ng/mL。对于涉及小样本的生物应用,如下 一代测序和 DNA 扩增产物的定量,需要更敏感的方法。来自 Thermo Fisher Scientific 公司的 Quant-iT PicoGreen dsDNA 检测试剂盒对 DNA 更特异,比传统光吸收方法的灵敏度高约 1000 倍。 如产品手册所述,此实验在微孔板格式中采用单 一染料浓度的动态范围从 250 pg/mL 到 1000 ng/mL。在这 里,我们证明了使 用 Molecular Devices 公司 SpectraMax® 酶标仪和 Quant-iT PicoGreen 实验,用户可以可靠地测量低 至 50 pg/mL 的双链 DNA 浓度。
为了最大化实验的灵敏度,需要使用最佳的激发和发射波长。 与基于滤光片的酶标仪不同,SpectraMax iD5 多功能酶标仪 和其他 SpectraMax 酶标仪中的双单色器允许在酶标仪规定 的范围内选择任何波长。确定为实验提供最佳灵敏度和动态 范围的激发和发射波长是很重要的,因为这些波长可能与基 于滤光片的酶标仪使用的波长以及试剂盒手册中推荐的波长 有所不同。
材料
- Quant-iT PicoGreen dsDNA 检 测 试 剂 盒 (Thermo Fisher Scientific cat. #P7589)
- 96- 孔全黑微孔板 (Greiner Bio-One cat. #655076)
- 避光黑色微管 (Argos cat. #T7100BK)
- 酶 标仪 ( 注 : 这 里 没 有 列 出 的 其 他 SpectraMax 酶 标 仪 对 PicoGreen 的实验具有类似的性能 )
- SpectraMax® iD5 多 功 能 酶 标 仪 (Molecular Devices cat. #iD5)
- SpectraMax® i3x 多 功 能 酶 标 仪 (Molecular Devices cat. #i3x)
- SpectraMax® M5 多 功 能 酶 标 仪 (Molecular Devices cat. #M5)
- SpectraMax® Gemini ™ EM 酶 标 仪 (Molecular Devices cat. #EM)
方法
仪器和方案设置
- 打开酶标仪。
- 启 动 SoftMax®Pro 软 件, 并 从 实 验 方 案 下 拉 菜 单 中 打 开 PicoGreen 荧光方案。根据您使用的 SpectraMax 酶标仪, 您可能需要输入检测的优化设置 ( 见表 1 )。
- • 通过点击“Settings”,在屏幕的左侧选项选择要读取的孔 和检测孔板类型。
- 点击模板按钮,打开一个窗口,您可以将微孔板的孔分配 到预先设置的模板组。使用下拉菜单选择适当的模板组。 PicoGreen 荧光方案中有预先设置的模板组,包括标准品、 未知样品和未知 _NoDiln( 用于未稀释的样品 )。将孔分配给 预设的模板组,用读取微孔板时获得的相应数据填充方案中 的组表。
实验准备
该方法遵循 QuantiT PicoGreen dsDNA 试剂和试剂盒产品信 息表的说明,除了实验体积从 2.0 mL 按比例减少到 200µL, 以适应 96 孔微孔板格式。
- 用无 DNA 酶的蒸馏水将试剂盒中的浓缩缓冲液稀释 20 倍, 制备成 1X TE 缓冲液 (10 mM Tris-HCl, 1 mM EDTA, pH 7.5)。
- 将浓缩 DMSO 溶液稀释 200 倍,在 TE 缓冲液中配制 QuantiT PicoGreen 试剂的工作水溶液 ( 如上所述 )。建议在塑料容 器中配制溶液,而不是在玻璃容器中,因为试剂可能会吸附 在玻璃表面。用棕色或黑色的试管,或用箔片覆盖以保护溶 液不受光。该溶液应在配制后的几个小时内使用。
- DNA 标准曲线 : 在 TE 中制备 2µg/mL 的 dsDNA 原液。试剂 盒提供的 lambda DNA 标准品可以用 TE 稀释 50 倍,制成 2 µg/mL 的溶液。
注 : 在某些情况下,最好使用与被测类型相似的 DNA 做标准 曲线。
Emission: 535 nm
bandwidth 9 nm
Emission: 525 nm,
bandwidth 15 nm
Emission: 525 nm,
Emission cutoff: 515 nm
Emission: 525 nm,
Emission cutoff: 515 nm
Integration time: 200 ms
Read height: Optimize for microplate used
Flashes per read: 10
Read height: Optimize for microplate used
Flashes per read: 10
Flashes per read: 10
***表 1 SpectraMax 酶标仪的仪器设置。*对于 SpectraMax iD5 和 i3x 酶标仪,读取高度设置应针对所使用的微孔板进行优化。注 : 列出了其他具有类似 性能的酶标仪。
- 根据需要,可以从 1 ng/mL 到 1 µg/mL 制备高范围的标准 曲线,也可以从 25 pg/mL 到 25 ng/mL 制备低范围的标准 曲线。对于高范围或低范围曲线,可以使用 1:10 的稀释。对 于低范围曲线,将 2 µg/mL 的溶液稀释 40 倍,得到 50 ng/ mL 的起始溶液。
为满足本应用要求,建立了 50 pg/mL 到 1 µg/mL 的一系 列标准品,稀释倍数为 1:3。 - • 以每孔 100 µL 将标准品移液至 96 孔全黑微孔板中,最好 重复三次。确保包含一组只有 TE ( 不含 DNA ) 的缓冲液空 白孔。
- • 每孔加入 Quant- iT PicoGreen 试剂工作水溶液 100µL( 这 会使标准品在孔中以 1:2 稀释 )。搅拌或用振板功能混匀并 在室温下避光孵育 2 - 5 分钟。
读板
- 如果使用的是 SpectraMax M 系列酶标仪,请确保紫色孔 板适配器在微孔板读取抽屉的托架上。把微孔板放在抽屉 里。
- 点击 SoftMax Pro 软件中的 Read 按钮。
分析数据
- 微孔板读取后,相对荧光单位 (RFUs) 将显示在孔板部分。 数据将在设置模板时创建的组表中进行分析。关于来自分组 表的代表性数据示例,请参见表 2。
- 模板中指定的标准品 ( 因此显示在标准组表中 ) 将自动绘制 在方案的标准曲线部分。
- 默认情况下应用线性曲线拟合,但是在绘制一个很宽的动态 范围内的标准曲线时,可以使用对数曲线拟合。从图形部分 的 Fit 下拉菜单中选择曲线拟合。
表 2 DNA 标准品。
结果
使用 Quant-iT PicoGreen dsDNA 检测试剂盒和 SpectraMax 酶 标 仪 检 测 50 pg/mL 至 1 µg/mL 的 DNA 标 准 品 ( 数 据 来 自 SpectraMax iD5 酶 标 仪,但 其他 SpectraMax 酶 标 仪 给 出类似的结果 )。SoftMax Pro 软件自动计算每组标准品重复 的平均 RFU,标准偏差和 %CV。在 SoftMax Pro 软件中使用 对数 - 对数曲线拟合绘制了一条标准曲线 ( 图 1 )。采用 96 孔 微孔板格式,检测限为空白样品标准偏差的 3 倍,灵敏度可低 至 50 pg/mL。这远远低于 Quant-iT PicoGreen 检测产品说 明书中规定的 250 pg/ mL 的下限。图 2 为高范围 (A) 和低范 围 (B) 的标准曲线。在标准品范围内线性良好 ( 图中各曲线的 r 2 ≥ 0.99 )。

图 1 DNA 标准曲线。 采用 Quant-iT PicoGreen dsDNA 检测试剂盒, 在 SpectraMax iD5 酶 标 仪 上 检 测 50 pg/mL 至 1000 ng/mL 范 围 内 的 DNA 标准品。采用 SoftMax Pro 软件中的对数 - 对数曲线拟合 (r2=1.00) 绘制标准曲线。

***图 2 高范围 (A) 和低范围 (B) 标准曲线。*使用 SoftMax Pro 软件中的对数 - 对数曲线拟合绘制曲线 ( 两条曲线均为 r2=0.99 )。
总结
Quant-iT PicoGreen dsDNA 检测试剂盒在 SoftMax Pro 软件的 SpectraMax 酶标仪上运行时,是一种快速、灵敏的双链 DNA 检测方法。该软件的分析功能以易于阅读、用户可自定义的报告格式提供定量。一个预先配置的方案在软件中可用,以方便快速 检测设置。