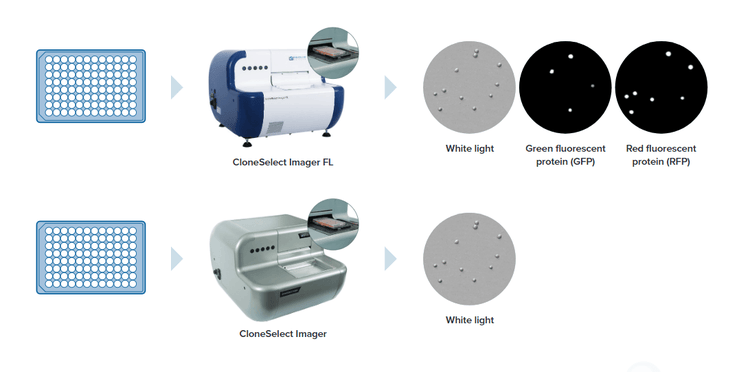
CloneSelect Imager (CSI and CSI FL)
High-speed fluorescence and white light imaging, intelligent data analysis, and monoclonality report generation
The CloneSelect Imager FL is a next-generation fluorescent imaging solution for the assurance of monoclonality and automated confluence across diverse cell types
Demonstrating that cell lines are monoclonal – or that a gene was edited as expected – can be a time-consuming and highly-subjective process when relying on conventional technologies. The CloneSelect® Imager and CloneSelect® Imager FL are a high-throughput automated solutions for imaging and analyzing mammalian cells. Tracking the formation of a colony from a single cell is effortless as barcoded plates are tracked over time. Automated acquisition and analysis provides accurate, objective, and consistent results.
With high-resolution white light imaging, the CloneSelect Imager provides automated confluence, monoclonality assurance and industry leading acquisition times with the ability to image a 96-well plate in under two minutes.
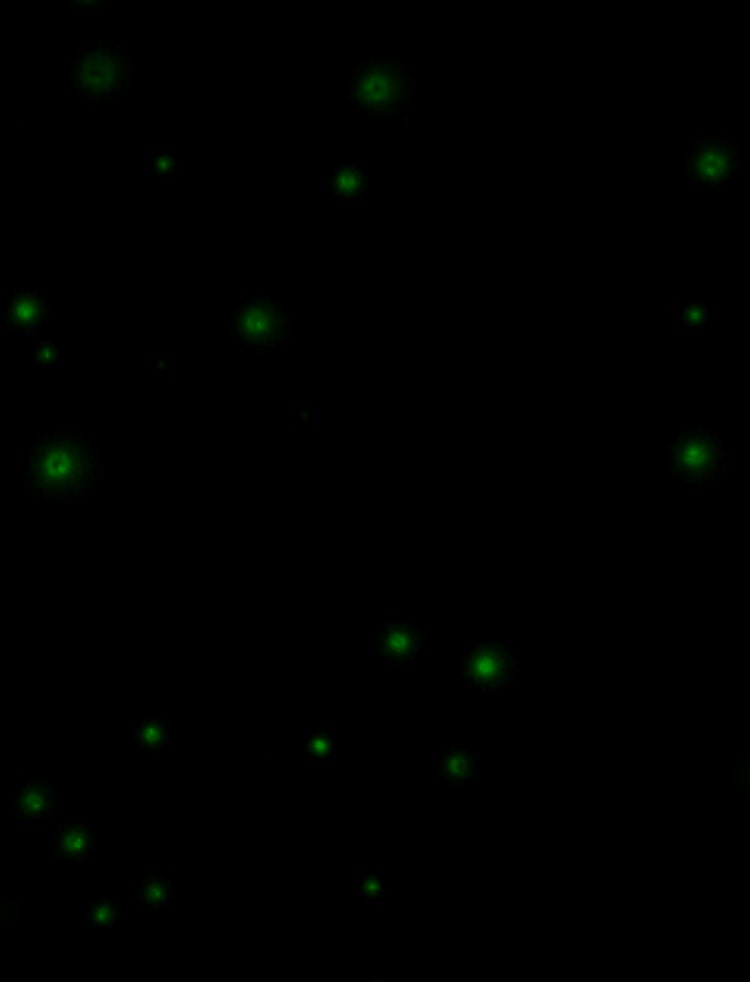
The all new CloneSelect Imager FL features high contrast multichannel fluorescent and white light imaging that allows for accurate single cell detection and proof of monoclonality at day 0. Streamline your workflow with comparative confluence assays to identify and verify gene edits.

Demonstrated IND success
The Monoclonality Report feature streamlines the creation of supporting documentation for regulatory agencies. Reports are automatically generated based on parameters you select. The Monoclonality Report is an audit-ready document that supports filing for an Investigational New Drug (IND) Application with the FDA. (21 CFR Part 312)

Multichannel imaging and automated confluence
Algorithms are optimized for accurate cell detection and address varying cell types and conditions. Publication-ready high-resolution imaging provides automatic confluence analysis and monoclonality assurance.

Rapid single cell confirmation
The imager delivers industry leading acquisition times, imaging a 96-well plate in as little as two minutes and can verify monoclonality from day zero with fluorescence.
Small footprint, high speed, powerful, imaging system for monoclonality verification
Features

White light and fluorescent imaging
Image every well in every plate with rapid acquisition times with label-free white light. Multichannel fluorescence imaging provides additional confidence of monoclonality and comparative confluence assays (red vs green).

Data tools to accelerate research timelines
The software automatically calculates confluence measurements and generates growth curves, heatmaps, and image montages. Measurements for every well are automatically tracked over time. Streamline multiple steps (imaging, sample tracking, data analysis and report generation) in the cell line development workflow for IND filing.

Quickly image a variety of plate formats and cell types
Image a 96-well plate in under two minutes. Compatible with adherent or settled suspension cell types such as CHO, HEK, hybridomas, iPSCs, and many other cell types.
Intelligent analysis with easy-to-use software
The software automatically calculates confluence for each imaging time point. Growth curve, image montage, total growth, and mean rate are generated automatically and are exportable. Guided software user interface allows for simple 1-hour training.

High-resolution images
High-speed fluorescence and high-resolution white light imaging with accurate detection of single cells including debris. Image visualization and data tracking over multiple days.

Custom automation options*
The Automation and Customization Team offers a variety of custom services from robotic plate loading to fully automated workstations with liquid handling and incubation. We have built out custom automated work cells with the CloneSelect Imager for iPSC workflows, full assay systems and drug toxicity and characterization research.
*Price, time to deliver and specifications will vary based on mutually agreed technical requirements. Solution requirements may cause adjustment to standard performance.
Streamlined Workflow
Image.
Analyze.
Report.
(1) Image
Seed one cell per well and image at any point
- Optimize clonal outgrowth - The system is particularly useful for optimizing clonal outgrowth strategies when platform approaches are not suitable e.g. when investigating new cell lines or variants
- Diverse cell types - Compatible with adherent or settled suspension cell types such as CHO, HEK, hybridomas, IPSCs, and many other cell types.
(2) Analyze
Assess cell growth objectively and quantitatively
Track and view growth of every cell line
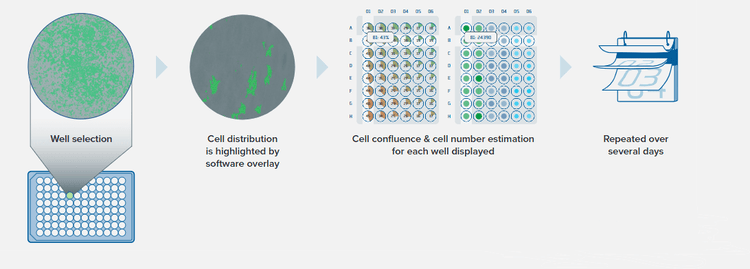
(2) Analyze
Growth curves calculated and displayed
Electronically track and store plate data: cell confluence, cell number estimation, and growth curve
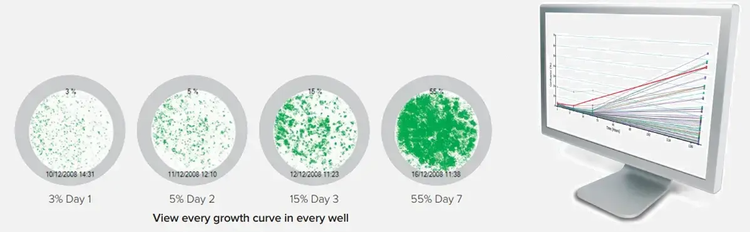
(3) Report
Verify monoclonality
After initial seeding, CloneSelect Imager system can image every well, at any time point, using a ‘loci of growth’ functionality to highlight those wells that contain a single colony.
- Focus on wells with a single loci of growth and view image history to verify monoclonality with the CloneSelect Imager using white light
- With the CloneSelect Imager FL automatically verify monoclonality from day zero with fluorescence

(3) Report
Monoclonality Report provides image evidence of monoclonal cell bodies
With a few simple clicks, the Monoclonality Report feature on the CloneSelect™ Imager (CSI) objectively organizes the supporting image evidence needed to establish clonality into an easily shareable report, saving researchers hours typically required to do the same process manually.
- Easily identify and select single-cell and artifact regions to include in report
- Export high-resolution images of single cells, artifacts, and entire well (optional)
- Automatically identify single cells with fluorescence version of CloneSelect Imager
- Publish monoclonality report in PDF or Word Format

(3) Report
Monoclonality Report feature automatically generates image-based data in a shareable format
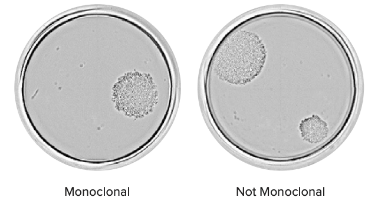
View entire well at final time point
Quickly determine clonality of a cell line by visually inspecting the presence of multiple colonies in a single well. For example, the well on the left shows one colony while the cell on the right shows two.
Monitor cell line development over time
To characterize the growth from a single cell to a colony, cell regions can be designated and adjusted for each time point in a series. The image bellow shows a single cell on Day 0 and two cells on Day 1 confirm monoclonality.
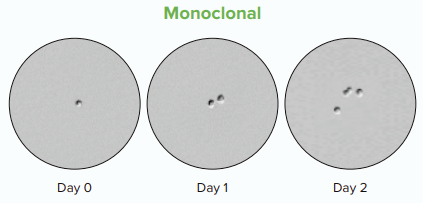
A single cell on Day 0 and two cells on Day 1 confirm monoclonality.
(3) Report
Monoclonality Report feature automatically generates image-based data in a shareable format
Divide an entire well into individual images
Export an entire well into 81 separate images to objectively confirm the absence of another cell. Below, a selected region is displayed over time.
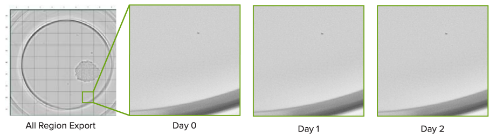
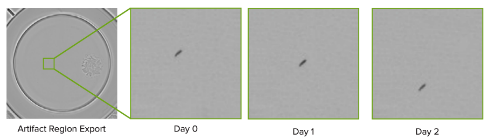
Highlight regions to review non-cell objects
Selectively highlight parts of a well to differentiate cells from ambiguous objects. A selected artifact region and its corresponding images over time is shown here.
FAQ
What are CloneSelect Imager (CSI) and CSI FL?
CloneSelect Imager (CSI) and CSI FL are advanced imaging systems designed for automated clone screening in mammalian cell culture. They provide high-throughput imaging and analysis capabilities to streamline the clone selection process.
What are the key features of CloneSelect Imager (CSI) and CSI FL?
CloneSelect Imager (CSI) and CSI FL offer several key features, including automated imaging and analysis, high-resolution imaging with brightfield and fluorescence capabilities, customizable imaging protocols, and integrated software for data management and analysis.
The stringent regulations of cell line development (monoclonal) can be easily met using the fluorescence detection capabilities of CSI-FL to confirm a single cell and to provide a Day O assurance of monoclonality.
How can CloneSelect Imager (CSI) and CSI FL optimize the clone screening process?
CloneSelect Imager (CSI) and CSI FL can optimize clone screening by automating the imaging and analysis steps, reducing manual labor, and increasing throughput. The systems provide accurate and consistent image capture, enabling the identification of high-quality clones based on desired characteristics.
The fluorescent method of detection rules out the detection of debris, dust, air bubbles and unwanted cells. It is an added advantage to our white light monoclonality verification, which screens high-value clones during the imaging process. Fluorescence to screen viable healthy cells could be achieved through our cell-specific fluorescent dye (Cell Tracker Green CMDFA) or a label-free approach i.e., expression of fluorescence protein as a marker via gene inserts.
What types of assays can be performed using CloneSelect Imager (CSI) and CSI FL?
CloneSelect Imager (CSI) and CSI FL support a wide range of assays, including viability and confluence measurements, cell counting, and fluorescence-based assays such as GFP expression analysis. The systems are versatile and can be adapted to specific experimental requirements.
The Fusion software in CSI-FL analyzes the cell counts and the presence of a fluorescence marker for the gene edit carried out. Each well is analyzed through a robust single-cell identification technique using both white light and fluorescence images.
Can CloneSelect Imager (CSI) and CSI FL integrate with existing laboratory workflows?
Yes, CloneSelect Imager (CSI) and CSI FL are designed to integrate seamlessly with existing laboratory workflows. The systems can be connected to robotic platforms for fully automated processes and can also export data to various formats for further analysis or integration with other software tools.
How does CloneSelect Imager (CSI) and CSI FL contribute to scientific research?
CloneSelect Imager (CSI) and CSI FL play a crucial role in scientific research by enabling efficient and reliable clone selection in mammalian cell culture. By automating the imaging and analysis processes, researchers can save time and resources, leading to faster and more accurate results.
Can CloneSelect Imager (CSI) and CSI FL be used in various laboratory workflows?
Absolutely! CloneSelect Imager (CSI) and CSI FL are versatile imaging systems that can be seamlessly integrated into a wide range of laboratory workflows. Whether you're working on monoclonal antibody production, cell line development and characterization, disease modeling or screening for high-throughput cell-based assays, these systems can streamline and optimize your workflow.
What advantages does CloneSelect Imager (CSI) and CSI FL offer over manual clone screening methods?
CloneSelect Imager (CSI) and CSI FL offer significant advantages over manual clone screening methods. These come with special software that successfully eliminates the subjectivity and variability associated with manual analysis, ensuring consistent and reproducible results. These results come with data reports, including various details, and image-based proofs - especially for the monoclonality verification. Additionally, the automated imaging capabilities significantly increase throughput and reduce the labor-intensive nature of clone screening and increase accuracy of data-driven assays
Can CloneSelect Imager (CSI) and CSI FL be used for analyzing cell viability and growth rates?
Yes, CloneSelect Imager (CSI) and CSI FL are equipped with advanced algorithms that allow the analysis of cell viability and growth rates. By capturing high-resolution images at multiple time points, researchers can accurately monitor cell growth dynamics, assess viability, and make informed decisions about clone selection.
Can CloneSelect Imager (CSI) and CSI FL help in optimizing the production of recombinant proteins?
Absolutely! CloneSelect Imager (CSI) and CSI FL are valuable tools for optimizing the production of recombinant proteins. By efficiently identifying high-producing clones based on fluorescence intensity or other parameters, researchers can save time and resources during the cell line development process. This enables the selection of clones with high expression levels and desirable protein characteristics, leading to improved protein production workflows.
Latest Resources
Featured Applications
Applications of CloneSelect Imager
Specifications & Options of CloneSelect Imager
Automation compatibility
API suite available for robotic integration. Please contact us for details
Download Brochure